I discovered something new this week — or rather, my computer did.
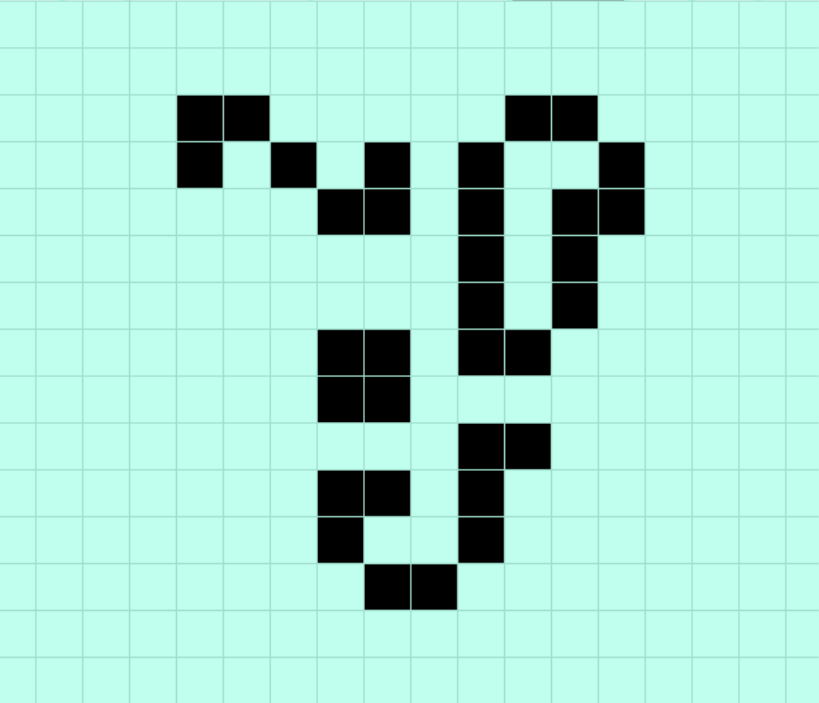
discovered by an automated run using APGSearch
Conway’s Life (which I’ve covered before) is a fascinating, simple yet Turing-complete set of rules to be run on a 2D binary lattice. Each generation, cells are born if they have exactly three live neighbors in the previous generation, and cells which were alive and had two or three neighbors stay alive. Otherwise, the cell dies (or no cell is created).
“Still Life” patterns are those Life patterns (like a 2×2 block of four) which are completely stable, and don’t change from one generation to the next. No new cells are generated, and the existing ones remain without any dying off. Absent any outside influence like a stray glider crashing into it, check back in a million generations and everything will still be where it is.
Still Life patterns are relatively common — start with any large random field and apply the Life rules and you’ll get lots of small ones — but larger ones don’t occur “naturally” (in Life sequences starting from random fields) nearly as often. The vast majority of patterns in Life aren’t stable, and large groups of randomly-initiated cells usually settle down into a mix of small Still Life patterns and small oscillators, along with gliders that shoot off in whichever diagonal direction they’re pointed in.
Life happens to be one of those interesting computational phenomena which is both easy to experiment with on a small scale and still interesting/challenging even for modern, fast computers. “Catagolue” [sic] is a project that has been underway since 2015 to try to catalogue all of the various patterns that exist in Conway’s Life. Programs like APGSearch can search through randomized primordial 16×16 “soup” patterns, evolving them according to the rules of Conway’s Life and running a quick analysis to see if anything interesting happens.
There are a huge number to go through, with 2^256 possibilities — but modern PCs are quick, going through tens of thousands of such patterns per second by running them in parallel. Every few minutes, the program uploads another “haul” of perhaps a few interesting finds, each time it checks ten million patterns. Contrast this with the way the “famous” Life patterns were found — by Conway and his graduate students, using tokens on a Go board! The glider — maybe the most famous Life pattern of all — was noticed when one student (Richard Guy) reported, “Hey, my bit is moving,” while helping Conway determine the evolution of the R-Pentomino “methuselah.”
Conway and his crew found most of the easy ones long ago. But we have cooler toys now. By simply running APGSearch in the background (for less than a day so far), I’ve already come across the Conway’s Life equivalent of a new, interesting beetle that we think nobody has come across before — the population-33 Still Life pattern shown above. (The same search also apparently provided the seventeenth confirmed sighting of a rare period-2 oscillator. Not nearly as exciting as a first find, but peer review is an important part of science, too, and it’s nice to see it applied so thoroughly here.)
I can’t wait to see what else it digs up.